GenCC FAQ Sections
Common Questions
What is the GenCC and why is it needed?
Several groups and resources provide information that pertains to the validity of gene-disease relationships; however, the standards and terminologies to define the evidence base for a gene’s role in disease are still evolving, and the community is in need of trusted and harmonized sources that define the level of evidence for a gene’s role in disease. To tackle this issue, the Gene Curation Coalition (GenCC) was formed.
The Gene Curation Coalition brings together groups engaged in the evaluation of gene-disease validity with a willingness to share data publicly, to develop consistent terminology for gene curation activities, and to facilitate the consistent assessment of genes that have been reported in association with disease.
What are the goals of the GenCC?
The GenCC is working to harmonize approaches to ensure gene-level resources are comparable and interoperable. This allows groups to work together most effectively and to provide consistent and useful resources for the community. The goals of the GenCC are as follows:
- Understand the aims, processes, information used, classification systems, and users of the different gene curation efforts
- Develop consistent terminology for validity assessment as well as inheritance, allelic requirement, and mechanism of disease
- Develop the GenCC Database, a shared resource to bring together gene-level resources
- Collaborate to harmonize gene-disease curations
What is gene-disease validity curation?
Gene-disease validity curation answers the question “Is a gene associated with disease?” A curation evaluates the evidence for a claim for a specific gene, disease, and mode of inheritance.
How do I cite/acknowledge the GenCC for database use?
The GenCC data are available free of restriction under a CC0 1.0 Universal (CC0 1.0) Public Domain Dedication. The GenCC requests that you give attribution to GenCC and the contributing sources whenever possible and appropriate. The accepted Flagship manuscript is now available from Genetics in Medicine (https://www.gimjournal.org/article/S1098-3600(22)00746-8/fulltext).
Example Attribution Statements:
The following curated content was obtained from the Gene Curation Coalition (www.thegencc.org) which includes contributions from the following organizations: Australian Genomics, ClinGen, ….. Gene-Disease Validity classifications [date accessed].
The authors would like to thank the Gene Curation Coalition (GenCC) for generating curated content used in this project. GenCC’s curated content was obtained at www.thegencc.org [date accessed] and includes contributions from the following organizations: Australian Genomics, ClinGen, ….
Citing the GenCC:
To cite a specific content on our website, please use the following format:
The Gene Curation Coalition. URL [date accessed].
Can I submit to the database?
The GenCC does not currently accept individual submissions but new groups can join per guidance below.
Can my group join the GenCC?
Groups seeking to join the GenCC must perform their own gene curations and 1) have content that the GenCC SC considers useful to the mission of GenCC; 2) be willing to share their curations publicly on the website; 3) adhere to ClinGen gene curation standards (Strande et al 2017), an equally rigorous framework, or provide a widely used existing public gene-level resource (e.g. OMIM, Orphanet); and 4) be able to use our standardized clinical validity terms and disease ontologies for their submissions. If you feel that your group meets this curation standard, please contact us to inquire about joining the GenCC.
Can I follow my favorite gene?
You cannot currently follow your favorite gene, but this feature will be available soon in a future release.
Can I download the data from this site?
Yes, there are download buttons available on multiple screens throughout the website. However, due to licensing restrictions, a GenCC download does not include OMIM data. OMIM data can be accessed and downloaded through https://www.omim.org/
Can I search using previous HGNC names or symbols?
You cannot currently search using old gene symbols, but this feature will be available soon. Please consult HGNC (www.genenames.org) to confirm that you are using the most current gene symbol for searches.
Are HGNC gene symbols and names stable?
HGNC are committed to making as few changes to gene symbols as possible (https://blog.genenames.org/Stable_Symbols), but some updates may still be necessary. See https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7494048/ for situations that merit a symbol update. To keep symbols stable, sometimes changes will be made to the gene name only.
Some submitters with large public resources have very small data sets. Why is this?
To test the process, some GenCC submitters have so far submitted only a small subset of their curation data. This data set will continue to expand over the coming months
Website Pages FAQ:
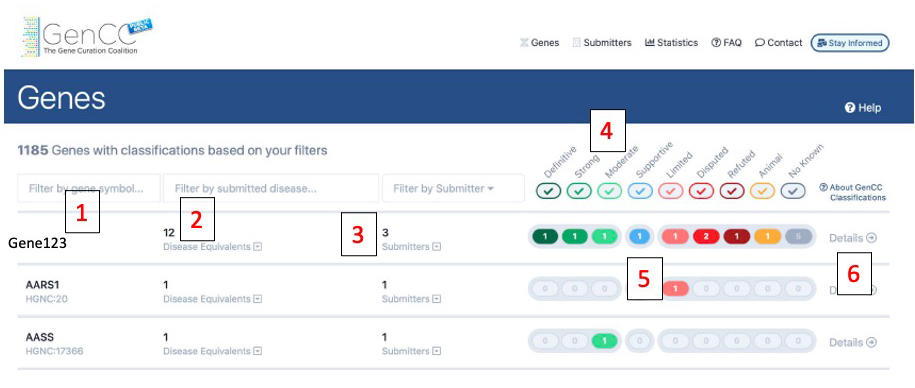
Landing Page
This is the landing page. All curated genes are listed. Numbers on the image correspond to descriptions below. All entries are collapsed but can be opened to see details. The detailed screenshot is found below:
1) This is the current approved HGNC gene symbol and its ID. Clicking the arrow next to it links you to the HGNC page for that gene. You can search and filter by HGNC gene symbol in the search box above the gene name.
2) GenCC maps all submitted disease terms (accepted ontologies are OMIM, Orpha, and MONDO) to the Monarch Disease Ontology (MONDO). Disease equivalents are the number of unique MONDO terms for which gene-disease associations are reported for the gene. Users can search by the submitted term in the search box above the disease equivalents.
3) This is the total number of unique submitters with classifications for the gene. Each submitter may have more than one classification. Users can filter by submitter at the top of the page.
4) All submissions are mapped to standardized clinical validity terms. The check boxes below the terms can be checked or unchecked to filter the data.
5) Each color corresponds to the standardized terms at the top of the page. The numbers represent the total submission with each evidence level.
6) Clicking on the details button will bring you to the gene-specific classification page.
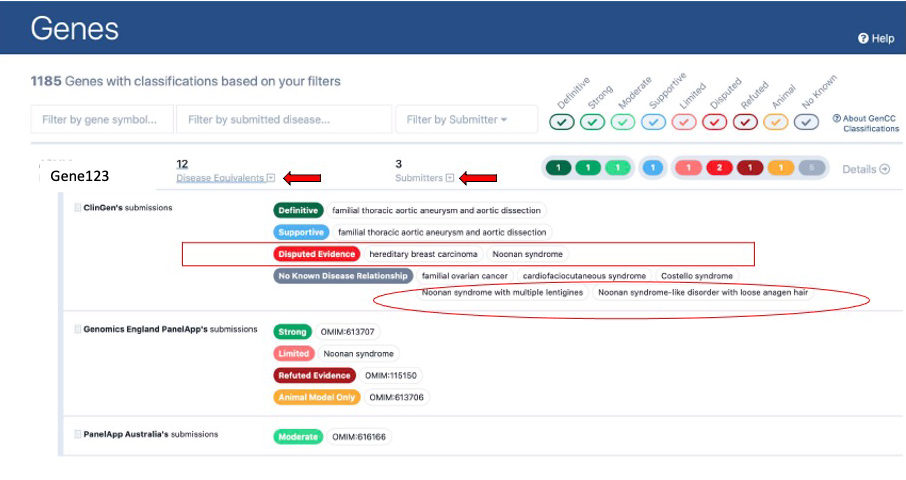
Landing Page Gene Features
The screenshot below is an expanded entry on the landing page (note: this is a fictitious example created to illustrate many features on one gene). Users can expand the entry by clicking on the arrow buttons next to “Disease equivalents” or “Submitters” (denoted by red arrows in the screenshot).
Each expanded entry is organized by submitter. Each line represents all submissions by a particular submitter with a certain evidence level. For example, boxed in red in this screenshot, ClinGen has two disputed evidence submissions (one for hereditary breast carcinoma and one for Noonan syndrome). When a submitter has too many submissions to fit on one line they will continue on to the next line, like ClinGen’s no known disease relationship submissions circled in red below.
When a user clicks the details button next to a gene entry, a gene page will open. Below are descriptive screenshots of the three tabs on the gene page: entries separated by classification, disease, and submitter.
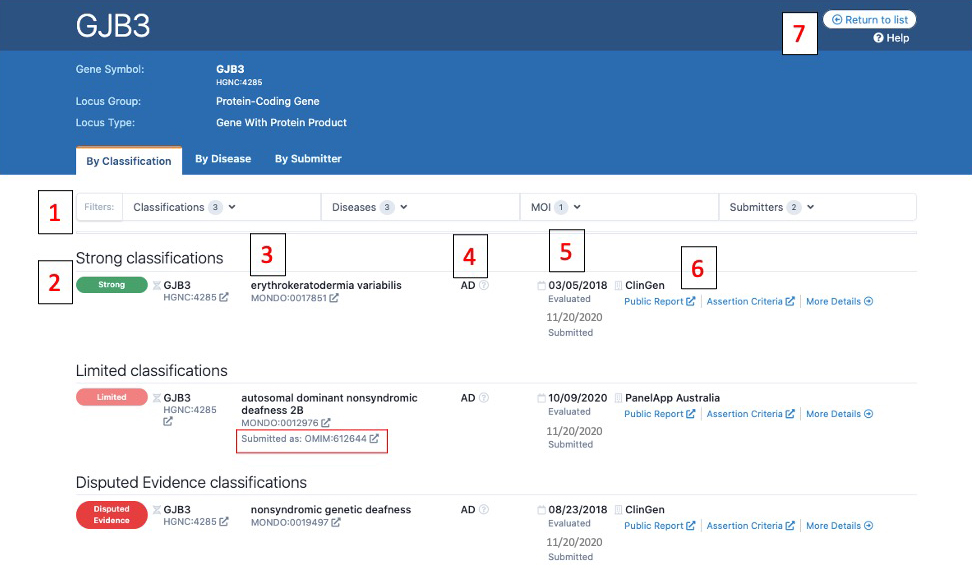
Gene Page By Classification
Above is a screenshot of a gene page where entries are displayed by classification. Numbers on the image correspond to descriptions below:
1) From these drop downs, users can filter the displayed list by Classification (e.g. only display Strong entries), Disease, Mode of Inheritance (MOI), or Submitter
2) The standardized clinical validity term is displayed with its corresponding color with the highest classifications listed first (e.g. Strong, then Moderate, then Limited)
3) All entries are mapped to Monarch Disease Ontology (MONDO) and the MONDO ID is displayed. If a submitter used a different ontology for submission (such as the PanelApp Australia classification), the original term or ID is displayed with “Submitted as:” (the OMIM ID boxed in red in the image)
4) The mode of inheritance.
5) The top date is the date the curation was evaluated. The bottom date listed in gray is the date that the curation was submitted to the GenCC.
6) This is the curation details section for each entry. The submitter is listed, along with an evidence summary, and all link outs (if available):
- Public report: public location of further curation information for this submission (website entry or publication)
- Assertion criteria: further information on the curation framework used for this entry
- More details: a link that will bring you to a specific page with more information about this group’s curation
7) The return to list button will take you back to the home page with the list of all curations.
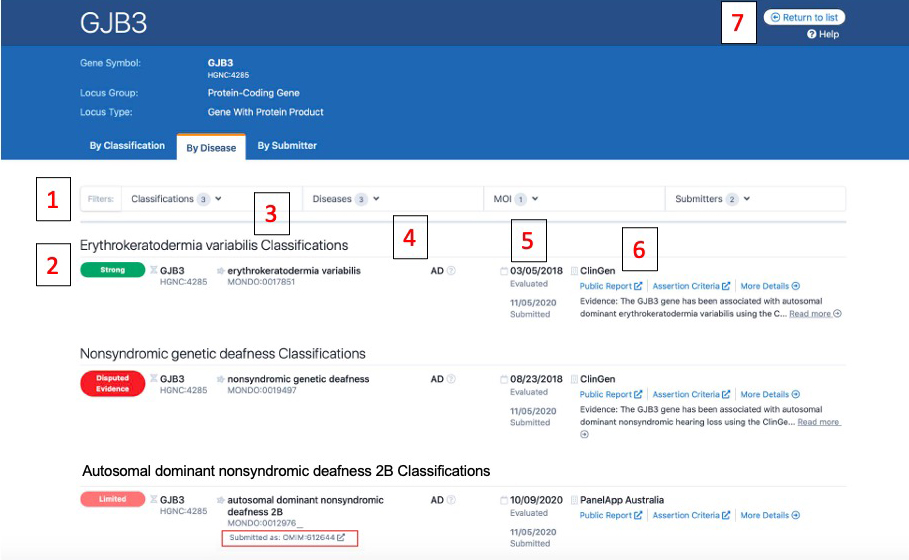
Gene Page By Disease
Above is a screenshot of a gene page where entries are displayed by disease. Numbers on the image correspond to descriptions below:
1) From these drop downs, users can filter the displayed list by Classification (e.g. only display Strong entries), Disease, Mode of Inheritance (MOI), or Submitter
2) The standardized clinical validity term is displayed with its corresponding color with the highest classifications listed first (e.g. Strong, then Moderate, then Limited)
3) All entries are mapped to Monarch Disease Ontology (MONDO) and the MONDO ID is displayed. If a submitter used a different ontology for submission (such as the PanelApp Australia classification), the original term or ID is displayed with “Submitted as:” (the OMIM ID boxed in red in the image)
4) The mode of inheritance.
5) The top date is the date the curation was evaluated. The bottom date listed in gray is the date that the curation was submitted to the GenCC.
6) This is the curation details section for each entry. The submitter is listed, along with an evidence summary, and all link outs (if available):
- Public report: public location of further curation information for this submission (website entry or publication)
- Assertion criteria: further information on the curation framework used for this entry
- More details: A link that will bring you to a specific page with more information about this group’s curation
7) The return to list button will take you back to the home page with the list of all curations.
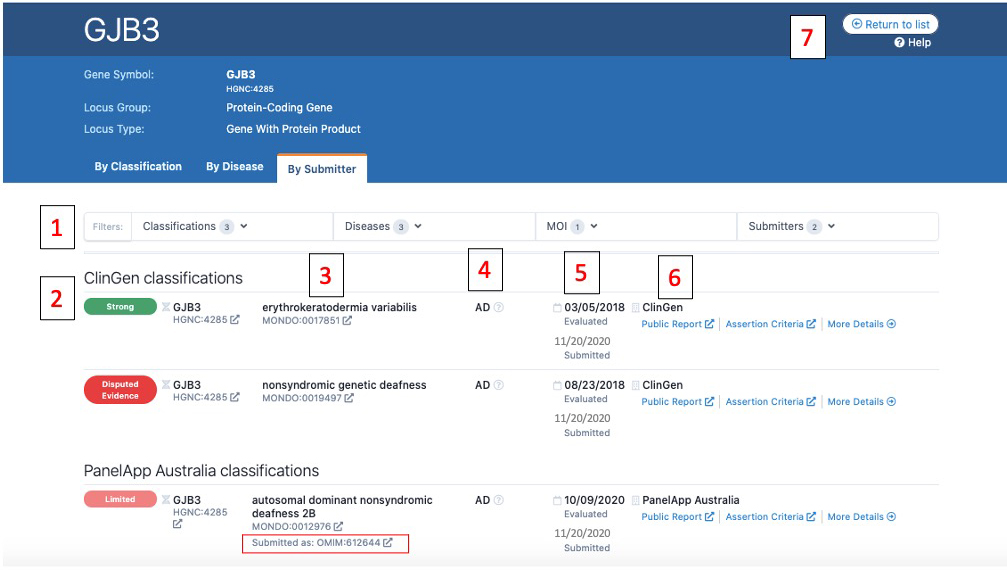
Gene Page By Submitter
Above is a screenshot of a gene page where entries are displayed by submitter. Numbers on the image correspond to descriptions below:
1) From these drop downs, users can filter the displayed list by Classification (e.g. only display Strong entries), Disease, Mode of Inheritance (MOI), or Submitter
2) The standardized clinical validity term is displayed with its corresponding color with the highest classifications listed first (e.g. Strong, then Moderate, then Limited)
3) All entries are mapped to Monarch Disease Ontology (MONDO) and the MONDO ID is displayed. If a submitter used a different ontology for submission (such as the PanelApp Australia classification), the original term or ID is displayed with “Submitted as:” (the OMIM ID boxed in red in the image)
4) The mode of inheritance.
5) The top date is the date the curation was evaluated. The bottom date listed in gray is the date that the curation was submitted to the GenCC.
6) This is the curation details section for each entry. The submitter is listed, along with an evidence summary, and all link outs (if available):
- Public report: public location of further curation information for this submission (website entry or publication)
- Assertion criteria: further information on the curation framework used for this entry
- More details: A link that will bring you to a specific page with more information about this group’s curation
7) The return to list button will take you back to the home page with the list of all curations.
Validity terms/Delphi Survey
To enable comparison of gene-disease validity curations across resources, we drafted harmonized definitions for differing levels of gene-disease clinical validity and then a Delphi survey approach was performed. In the first round, members of the GenCC took the survey and chose from term sets already in use by current efforts or suggested new terms. In the second round, all previous and suggested terms were incorporated, and the survey was sent out to the extended staff and members of each of the GenCC groups. In the third round, the survey was then sent to the larger international genetics community (e.g. American College of Medical Genetics Membership, American Society of Human Genetics Membership, the Global Alliance for Genomic Health Membership) with a 10 minute introductory video for context. Responses from the community were used to further narrow the term list, and the final harmonized term set with definitions are provided below. This set is used to map all other terms used by each curation effort participating in the GenCC and displayed on this curation site. Mapping exceptions include situations where a resource did not curate to the same level of granularity as the harmonized list and therefore a broader category of “Supportive” was used to represent a basic level of evidence for gene-disease association such as that used by OMIM and Orphanet.
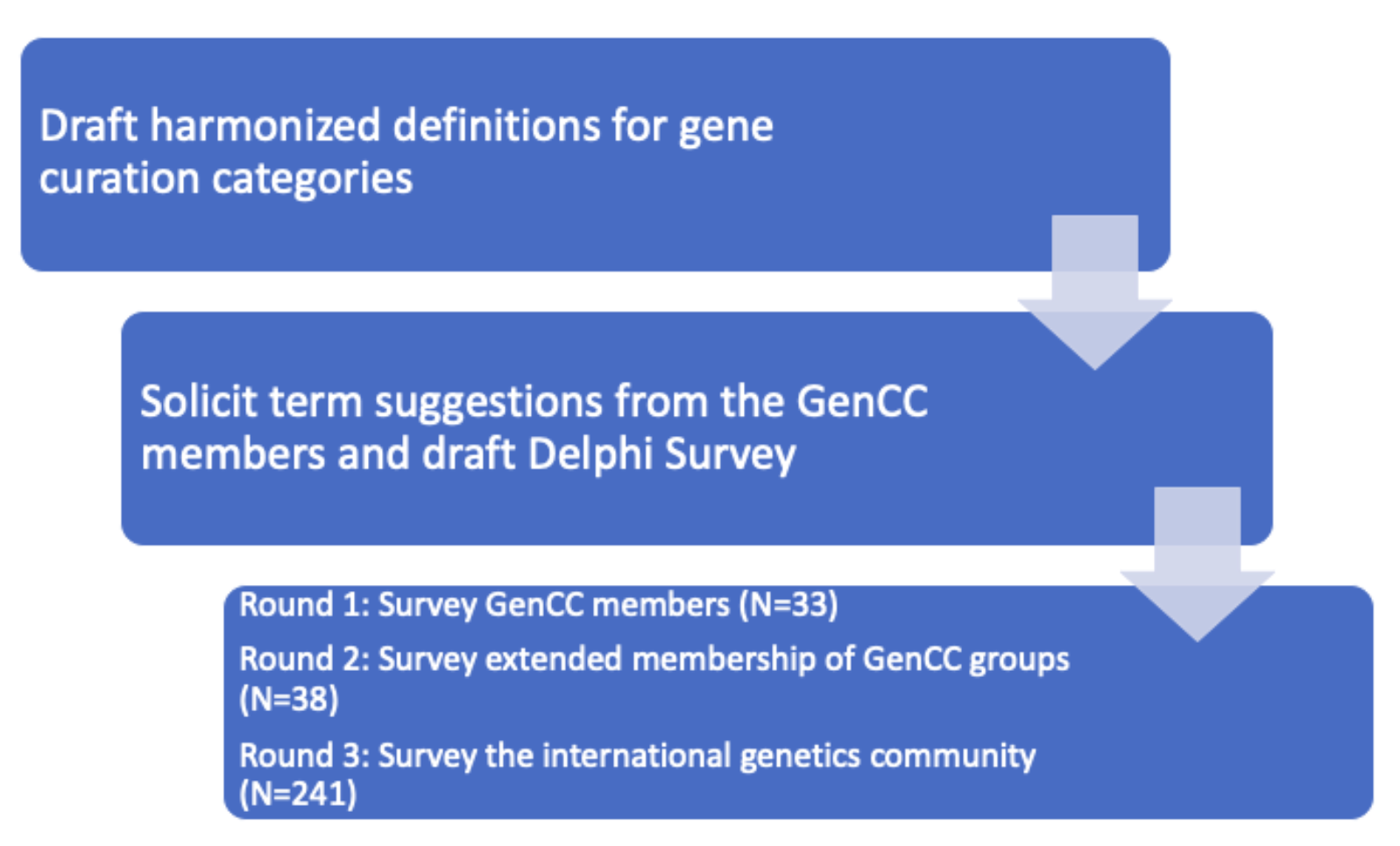 Preferred terms set and definitions*:
Preferred terms set and definitions*:
*This framework is intended to address highly penetrant monogenic forms of disease
Definitive
The role of this gene in this particular disease has been repeatedly demonstrated in both the research and clinical diagnostic settings, and has been upheld over time (at least 2 independent publications over 3 years’ time). No convincing evidence has emerged that contradicts the role of the gene in the specified disease. (Please note that not all submitters distinguish between strong and definitive. Thus, a strong submission may be the highest validity from a particular submitter. Please contact individual submitters if you have questions about this.)
Strong
The role of this gene as a monogenic cause of disease has been repeatedly and independently demonstrated providing very strong convincing evidence in humans and no conflicting evidence for this gene’s role in this disease.
Moderate
There is moderate evidence in humans to support a causal role for this gene in this disease with no contradictory evidence. The body of evidence is not large (e.g. possibly only one key paper) but appears convincing enough that the gene-disease pair is likely to be validated with additional evidence in the near future.
Limited
Little human evidence exists to support a causal role for this gene in this disease, but not all evidence has been refuted. For example, there may be a collection of rare missense variants in humans but without convincing functional impact, segregation data that could either arise by chance (e.g. across one or two meioses) or does not implicate a single gene, or functional data without direct recapitulation of the phenotype. Overall, the body of evidence does not meet contemporary criteria for claiming a valid association with disease. The majority are probably false associations.
Disputed
Although evidence has been reported, other evidence of equal weight disputes the claim.
Animal Model Only
No (or very little) human disease evidence exists, but a convincing animal model exists.
Refuted
There has been an assertion of a gene-disease association in the literature, but new valid evidence has arisen that refutes the entire original body of evidence.
No known disease relationship
No disease claim in any organism has ever been made.
